Projects
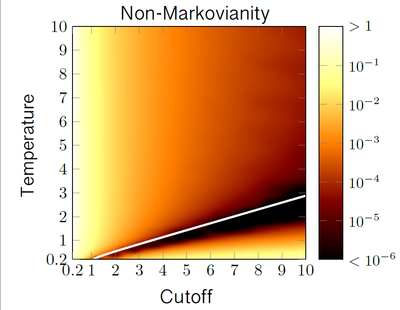
P1 - Classical and quantum non-Markovianity (Heinz Peter Breuer & Tanja Schilling)
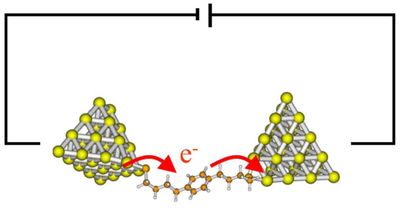
P2 - Charge transport in nanostructures: Current-induced forces and electronic friction (Michael Thoss, Thorsten Koslowski & Gerhard Stock)
P3 - Protein charge transfer away from equilibrium (Thorsten Koslowski & Michael Thoss)
Electron transfer in proteins is an essential biochemical reaction that can be found on all branches of the tree of life. In biological systems, charge carriers become trapped and undergo hopping transport between cofactors as the centers of excess charge localization.
Reducing the interactions in the complex aggregates of proteins and cofactors to few-site models that can be treated using methods from theoretical physics and chemistry is one of the central problems of biological charge transfer.
As part of the research unit 'Reducing complexity of nonequilibrium systems', we aim at computing and understanding electron transport through proteins under nonequilibrium conditions such as external fields and sources of charge. Here, transport through membranes and nanostructures usually occurs far from equilibrium and beyond the linear response regime. We plan to reduce the complexity of the biopolymers to simple models using dielectric theory and atomistic simulations. The resulting models will be addressed by a combination of Kirchhoff's equations for the voltages and currents and Master equations for the charges. The classical transport methods will be continuously validated employing high-level quantum transport approaches.
P4 - Non-Equilibrium Work And Dissipation in Coarse-Grained Models (Tanja Schilling & Lars Pastewka)
If one intends to coarse-grain the dynamics of a system that is out of thermal equilibrium, one needs to integrate out a non-stationary distribution of microstates. This implies that the entropy corresponding to this distribution will change with time, which in turn implies that there is a non-trivial contribution to the heat dissipated during processes performed by the coarse-grained variable. Our project will focus on this issue. Using a time-dependent projection operator formalism we will analyze the heat dissipated in a driven system.
We will first tackle this problem from the fundamental point of view of theoretical physics and then apply our findings to several numerical test-cases: microrheology of a polymer melt, sliding friction of solids on solids and the dissociation of NaCl in water. Finally we will develop a scale-bridging tool for molecular dynamics simulation that takes non-stationary memory effects into account.
P5 - Data-driven Langevin modeling of nonequilibrium processes (Gerhard Stock & Joachim Dzubiella)
Classical molecular dynamics (MD) simulations account for the structure and dynamics of molecular systems in microscopic detail, but become cumbersome with increasing system size. To drastically reduce the complexity, it is therefore often desirable to represent the considered process in terms of a small set of collective variables x(t). Employing the Langevin equation formalism, we may describe the stochastic motion of the system along x(t) in presence of the environment. While Langevin theory is well established, a relatively little explored aspect concerns its generalization to nonstationary processes. For one, we may consider an externally driven system, as is the case, e.g., in atomic force microscopy experiments, where we mechanically pull a molecule apart. On the other hand, we may start in a nonstationary state (e.g., resulting from a laser-induced temperature-jump or photoexcitation) and study the relaxation of the (otherwise not driven) system into its equilibrium state.
Adopting a Markovian Langevin formulation, in this work we are concerned with the situation that we have nonstationary data x(t) (provided by a time-dependent experiment or from an MD simulation) for which we want to construct a dynamical model that reproduces its statistical properties. To calculate multidimensional Langevin fields from given MD data, we have developed a data-driven Langevin equation approach, which we will extend to the nonequilibrium regime. In particular, we will identify appropriate collective variables that facilitate a Markovian description and outline ways to efficiently calculate nonstationary Langevin fields.
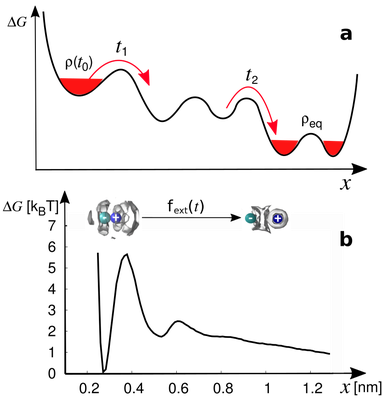
Modeling non-Markovian data using Markov state and Langevin models models
B. Lickert and G. Stock. J. Chem Phys. 153, 244112 (2020)
P6 - Feedback-controlled molecular transport through responsive polymer membranes (Joachim Dzubiella & Tanja Schilling)
Project P6 “Feedback-controlled molecular transport through responsive polymer membranes” by Dzubiella and Schilling is concerned with particle transport through dense polymeric membranes in nonequilibrium, which is relevant for applications like nano-filtration and desalination or medical treatments such as dialysis or selective drug delivery. Modern materials used for this purpose, such as thermosensitive polymers, often display a complex non-linear response behavior, in particular close to their critical solution (or ’volume transition’) temperature. Close to this temperature, the polymer network and therefore its permeability will respond to the inhomogeneous particle density distributions and feed back to the particle fluxes and transport in a highly nonlinear way in both space and time domains. In this project, this nonequilibrium and feedback-controlled transport of solutes driven through responsive polymer membranes will be studied at various levels of coarse-graining, ranging from ’microscopic’, i.e., monomer-resolved computer simulations of the cross-linked polymer network to reduced, low-dimensional Langevin and Fokker-Planck equations involving coarse-grained collective variables, to finally arrive at a one-dimensional generalized Langevin description with a memory kernel. Various nonequilibrium situations will be studied, from constant driving fields to full nonequilibrium stimuli situations. The dissipation-corrected targeted MD method developed in this RU will be employed for gaining insight into microscopic sources of friction appearing in the polymer network. Combining method developments to study coarse-grained nonlinear transport phenomena with highly relevant technical applications, P6 connects to the fundamental theoretical methods developed in this RU and applies and extends them to a “real life” problem of soft-matter science.
Figure: Left: Snapshot of monomer-resolved computer simulations of penetrants driven through a polymer network. Middle: Membrane mapped on energy and diffusion landscapes for coarse-grained theories, such as the Soluchowski equations or Brownian dynamics equations. Right: Resulting inhomogeneous penetrant density profiles (comparison of theory, dashed lines, versus simulations, solid lines. The curves in the latter example are for only little responsive polymers, hence the agreement for the profiles (and fluxes) is quite good.
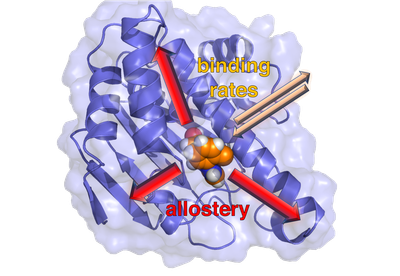
P7 - Nonequilibrium functional dynamics of proteins (Gerhard Stock & Steffen Wolf)
Biological processes in living organisms occur under inherent nonequilibrium conditions caused by local concentration changes of substrates such as ATP, ions and ligands. For single proteins, nonequilibrium reactions can be viewed as perturbations of a local equilibrium, followed by relaxation into a new local equilibrium.
One example for such processes is unbinding of ligands from host proteins due to ligand depletion within the blood stream. The prediction of binding and unbinding kinetics has become a recent focus of theoretical method development, as such kinetics directly affect the efficacy of a drug.
Another process of interest is the protein's own response to ligand binding or unbinding, which often amounts to a structural rearrangement of the biomolecule. A prime example are ligand-induced conformation transitions that give rise to allostery, that is, long-range communication between distant protein sites. In spite of its importance as elementary process of cell signaling as well as target in pharmaceutical research, there is surprisingly little known about the nonequilibrium aspects of allosteric communication.
The goal of this project is to gain insight into the nonequilibrium aspects of protein-ligand binding and the following allosteric communication. Since the conformational change depends on the binding mode of the ligand and conformational transitions may already occur during ligand (un)binding, both topics are closely related. Moreover, both approaches involve an enhanced sampling of rare events. To overcome present limitations of MD simulations and to make them applicable to the above described nonequilibrium processes, we will mainly pursue two approaches: dissipation-corrected targeted MD (dcTMD) to describe the binding or unbinding of ligands to proteins, and coarse-grained Markov modeling to describe the subsequent conformational change of the protein.
P8 - Viscous friction in mixed and boundary lubrication: Fluid dynamics far from equilibrium (Kerstin Falk, Steffen Wolf & Michael Moseler)
Project P8 is concerned with the crossover from the Newtonian fluid to nonlinear lubrication regimes. Friction in real tribological contacts is often determined by a mixed regime in-between dry sliding and hydrodynamic lubrication, where extreme local pressure reduces the lubricant film to a few molecular layers. In this regime, high shear rates can lead to a nonlinear response of the fluid, and confinement can lead to the break-down of hydrodynamics. The main goal of this project is to identify deviations from equilibrium dynamics due to both types of external constraints (confinement and imposed shear rate) and to formulate viscosity scaling laws for the non-linear regime, which can then be used for continuum modeling. To this aim, the lubricants viscosity, diffusion, and static and dynamic structure properties will be examined in nonequilibrium MD simulations under the relevant tribological conditions. In addition, the dcTMD method advanced in P7 will be applied to yield a time-resolved microscopic friction force exerted on individual lubricant molecules during diffusive events. Based on the results, a recently proposed diffusion and viscosity models for high pressure alkane liquids will be extended to anisotropic fluid structures. Methods developed within project P4 will yield a complementary mesoscopic description, which can be used to study the dynamics under a wider range of conditions than with fully atomistic MD. Moreover, the project will benefit from close collaboration with project P6 on solute dynamics in polymer networks and project P9 considering dry sliding friction.
K. Falk, D. Savio, and M. Moseler, Nonempirical free volume viscosity model for alkane lubricants under severe pressures, Phys. Rev. Lett. 124, 105501 (2020).
P9 - Energy dissipation in dry sliding friction (Michael Moseler & Lars Pastewka)
The goal of project P9 is a systematic derivation of the empirical Prandtl model, that is widely used to interpret solid-solid friction experiments. This systematic derivation will be based on atomistic simulations, lattice dynamics calculations, and - in collaboration with project P4 - projection operator techniques. Using a harmonic analysis to compute the deformation of an elastic solid during sliding, the effective dissipation felt by the sliding indenter will be calculated, which is used to systematically identify and remove fast degrees of freedom from the description. Using this controlled coarse graining of atomistic models, it will be possible to extract microscopically defined model parameters, check the strength and limitations of the model, and suggest generalized equations of motion in case the ordinary Prandtl model fails to reproduce MD reference simulations. Moreover, microscopic dissipation channels of the system can be identified and the systematically coarse-grained description will allow modelling long time scales, matching typical tribological experiments.
Results from project P9 feed into projects P4 and P5, and it complements the studies of friction with lubrication of project P8.

